Deamidation
Reduce the Risk of Charge Variant Formation via Deamidation
Understanding the events and mechanisms that lead to deamidation of a therapeutic protein reduce the risk of withdrawal later in drug development. ProteinMentor provides a comprehensive assessment of the extent of deamidation under varying conditions. This is achieved in real time for the full-length protein with little to no sample preparation. The platform can also be deployed for rapid assessment of the current batch versus a reference material within commercial operations.
ProteinMentor and Deamidation
Learn how ProteinMentor can advance your deamidation analysis
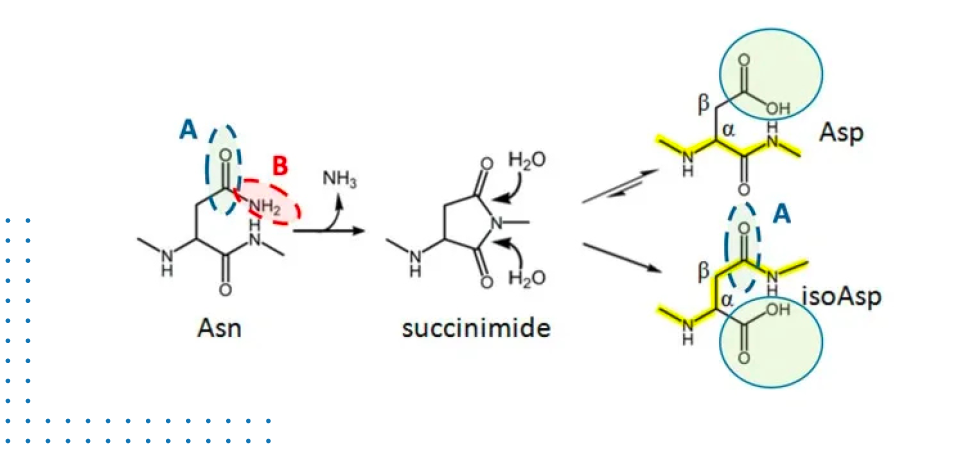
MECHANISM OF DEAMIDATION
The deamidation mechanism is well understood, and it is similar for both asparagine and glutamine residues. It is a non-enzymatic process that is induced by exposure to thermal and/or high or low pH stress. Deamidation results in the introduction of a negative charge where there formerly was none, giving rise to an acidic ‘charge variant’. Identifying the pathways that lead to deamidation is important to understand how to manage its prevention. Through clearer understanding of the process, steps can be taken within process development and formulation to reduce the risk factors that lead to deamidation.

EXTENT OF DEAMIDATION
The methods most commonly deployed for deamidation analysis today provide a static view in time. They typically compare different stressed samples with a control or ideal state of the drug substance. The methods generally also require buffer exchange and other extensive sample preparation steps prior to analysis. Often, they do not offer the ability to perform the analysis on the drug product in its formulation condition.
Using ProteinMentor, a comprehensive assessment of the extent of deamidation under varying conditions can be achieved in real time for the full-length protein. Which asparagine or glutamine residues are affected, and to what extent, can be determined, thus streamlining the process of evaluation.
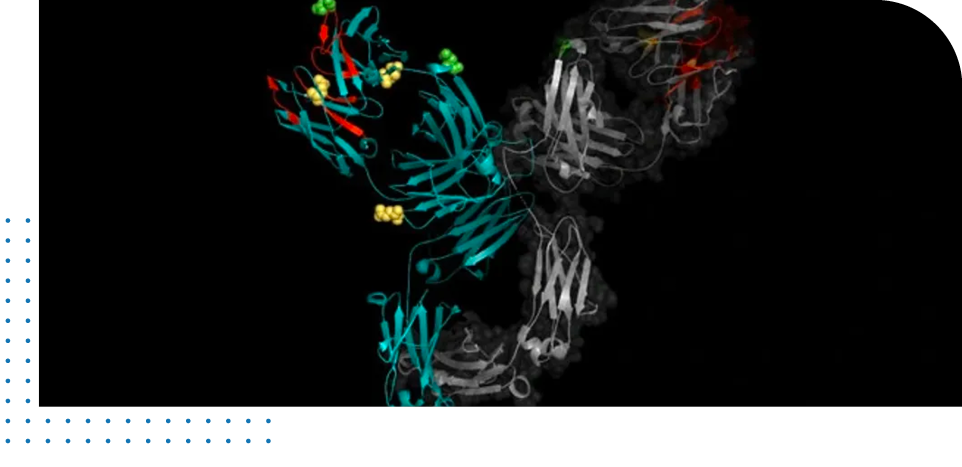
SITE OF DEAMIDATION
The empirical determination of the site of deamidation allows for risk assessment. If the evaluation occurs during the R&D phase of drug development then two options are available: (1) re-engineering of the identified hot-spot and/or (2) formulation optimization of the therapeutic candidate. The determination of the potential site of deamidation requires the amino acid sequence of the protein. The analysis using ProteinMentor is based on the assumption that neighboring residues are being perturbed by the deamidation event due to the introduction of a negative charge. Furthermore, the high-resolution structure is used to confirm solvent accessibility for the hot-spot, supporting the deamidation potential of the specific asparagine or glutamine residue.
IMPACT OF DEAMIDATION
If the deamidation site involves the complementarity-determining regions (CDRs), then the potential exists to impact efficacy. However, deamidation can have many implications on a therapeutic protein, from decreased domain stability, to self-association or aggregation leading to decreased half-life and safety concerns. Also, the potential of truncation of the therapeutic protein may exist. The cumulative potential risks associated with deamidation may result in lower production yields, as well as decreased safety and efficacy of the drug product that extends to storage and delivery.
Our approach provides a simplified workflow and real-time quantitative assessment for both asparagine and glutamine deamidation for the full-length therapeutic protein in the desired formulation.

Want to learn more?
Sign up to watch a webinar by Dr. Belinda Pastrana

Download the Deamidation Application Note
Follow the link to access the app note
